Software Project
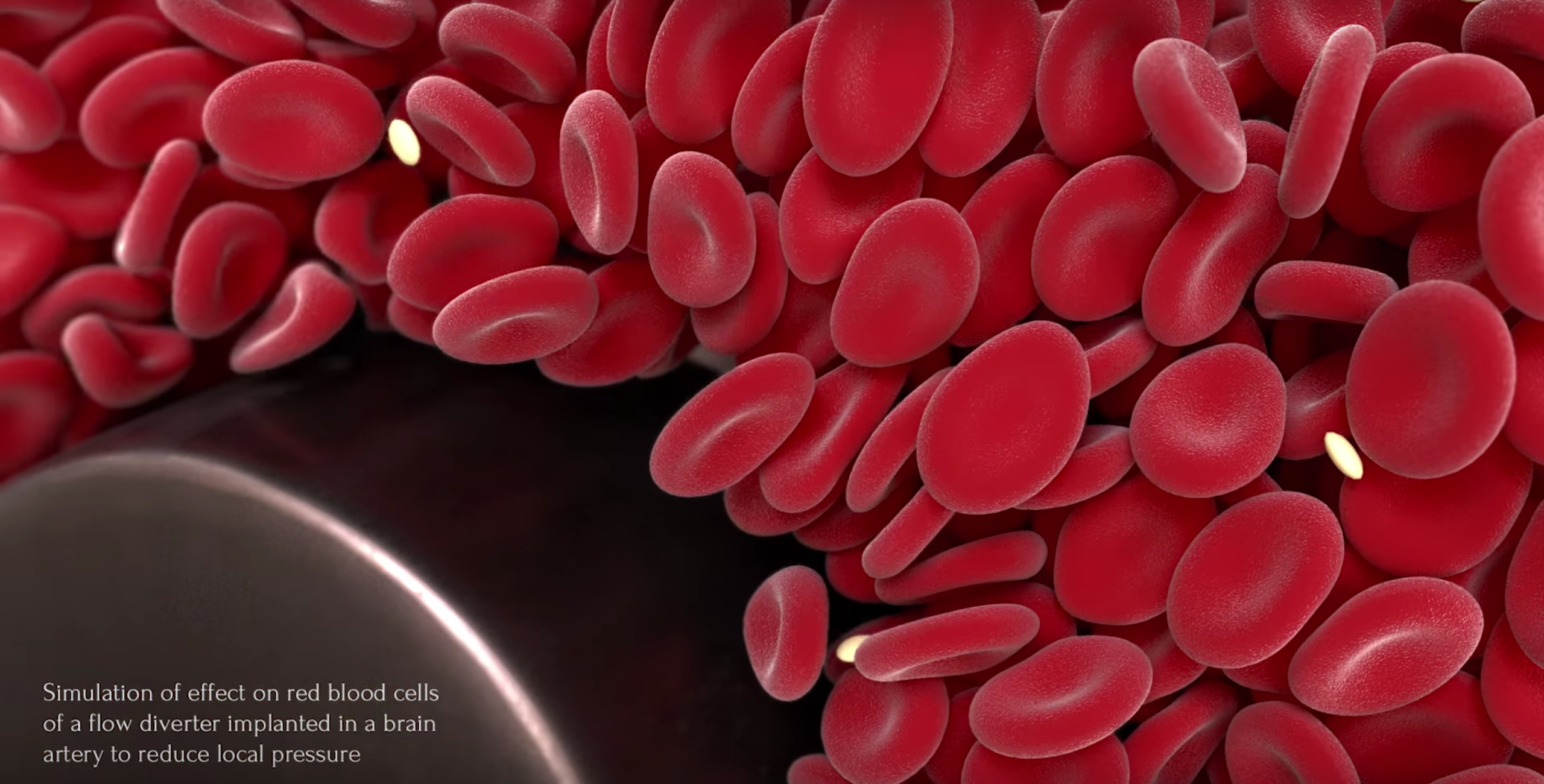
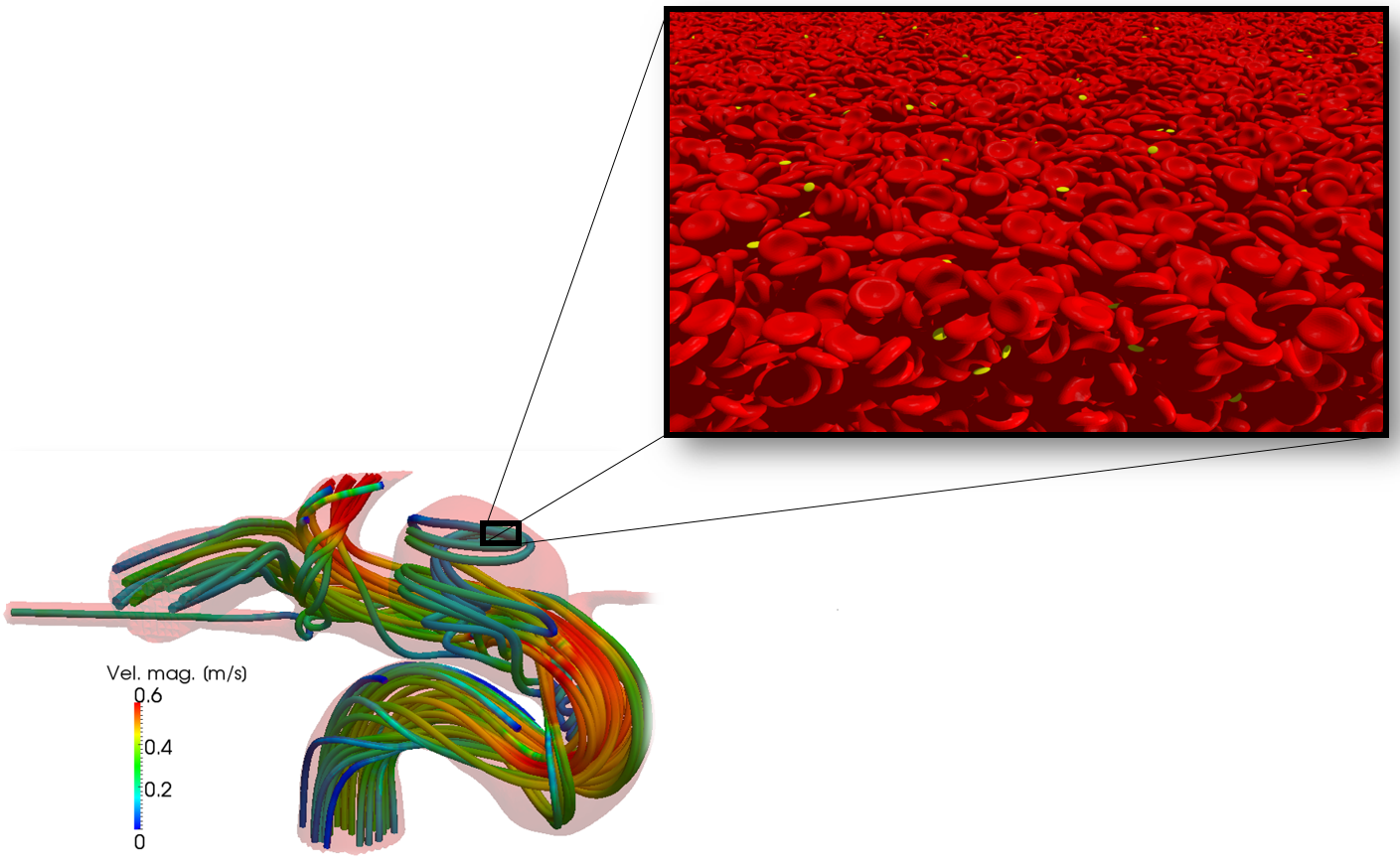
HemoCell, developed by the team of Prof Alfons Hoekstra at the University of Amsterdam (NL), is a high-performance library to simulate the transport properties of dense cellular suspensions, such as blood. It contains validated material model for red blood cells and additional support for further cell types (white blood cells, platelets). The blood plasma is represented as a continuous fluid simulated with an open-source Lattice Boltzmann Method (LBM) solver. The cells are represented as Discrete Element Method (DEM) membranes coupled to the plasma flow through a tested in-house immersedboundary implementation. HemoCell is computationally capable of handling a large domain size with a high number of cells (> 10^4-10^6 cells). The code is currently installed and optimised for Cartesius, Lisa, and SuperMUC (Leibniz Supercomputing Centre system), and can be used by anyone with a valid account and CPU allocation on any of these systems.
Offered By
Use scenario
Clinical research, Clinical decision support, In silico clinical trial
HPC motivation
Solve unreducible model; Multiscale model; Strongly coupled model.
Relevant links
- Hemocell official website
- Hemocell documentation
- Hemocell GitHub
- Computational Hemodynamics: From a notebook to HPC
- CompBioMed Webinar Introduction to Biomedical Applications on High Performance Computers
- CompBioMed Virtual Humans Film
Related Articles
- Závodszky G, van Rooij B, Azizi V, Hoekstra A. Cellular level in-silico modeling of blood rheology with an improved material model for red blood cells. Front Physiol. 2017;8(AUG):563.
 |
 |

