Software Project
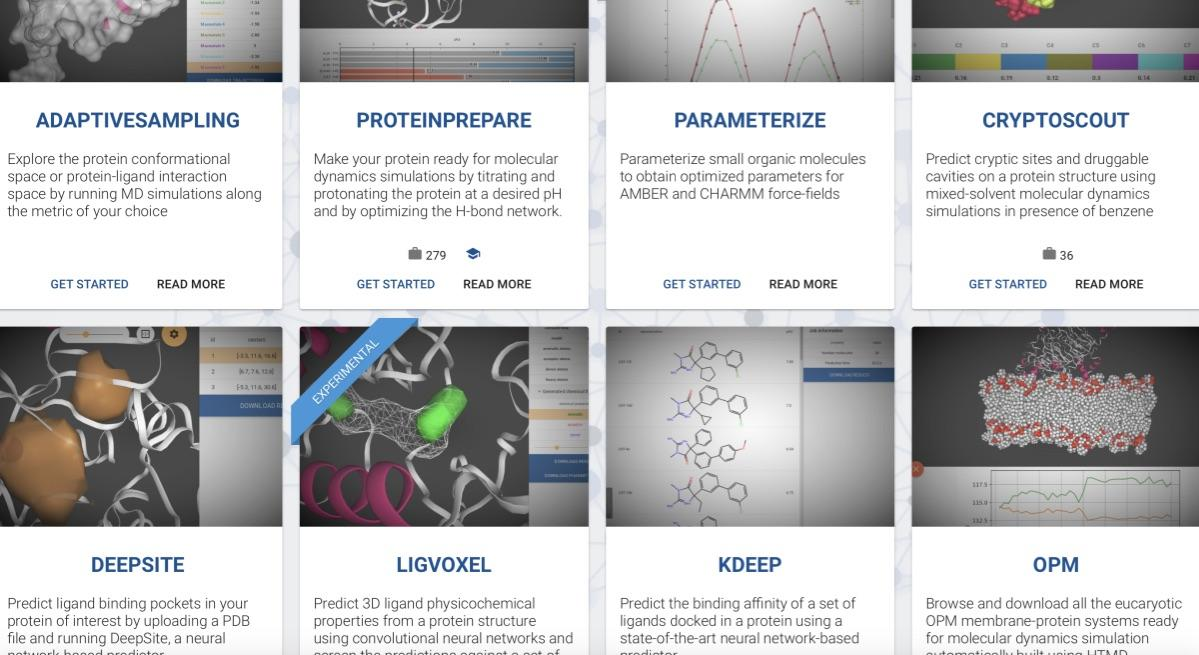
Playmolecule, developed by the team of Prof Gianni de Fabritiis at the Universitat Pompeu Fabra (ES), is an intuitive platform to access a diverse set of web applications for molecular research. It is a repository of free best-in-kind applications with a diverse set of solutions like molecular predictors and modelling tools. Simulations are run on GPUGRID for free or via Amazon AWS; The scalability is provided by Amazon via acecloud, the cloud interfacing software by Acellera.
Offered By
Use scenario
Non-clinical research; Drug discovery; Design & optimisation.
HPC motivation
Solve unreducible model; Do uncertainty quantification.
Relevant links
- PlayMolecule official website
- Accellera Youtube channel
- CompBioMed webinar: High Throughput Molecular Dynamics for Drug Discovery
- High Throughput Molecular Dynamics Simulations for Drug Discovery
Related Articles
- José Jiménez Luna, Miha Skalic, Gerard Martinez-Rosell, and Gianni De Fabritiis. KDEEP: Protein-ligand absolute binding affinity prediction via 3D-convolutional neural networks. Journal of Chemical Information and Modeling. (2018) DOI: 10.1021/acs.jcim.7b00650
- G. Martinez-Rosell, T. Giorgino, G. de Fabritiis. PlayMolecule ProteinPrepare: a web application for protein preparation for molecular dynamics simulations in J. Chem. Inf. Model 2017, June 10.
 |
 |


